
精神分裂症破坏蛋白1(DISC1)重组蛋白
产品名称: 精神分裂症破坏蛋白1(DISC1)重组蛋白
英文名称: Recombinant Disrupted In Schizophrenia 1 (DISC1)
产品编号: YB265Ra012
产品价格: 0
产品产地: 中国/美国
品牌商标: 钰博生物/Ybscience
更新时间: 2023-08-17T10:29:50
使用范围: null
- 联系人 : 陈环环
- 地址 : 上海市沪闵路6088号龙之梦大厦8楼806室
- 邮编 : 200612
- 所在区域 : 上海
- 电话 : 183****2235 点击查看
- 传真 : 点击查看
- 邮箱 : shybio@126.com
- 二维码 : 点击查看
精神分裂症破坏蛋白1(DISC1)重组蛋白
YB265Ra012
Disrupted In Schizophrenia 1 (DISC1)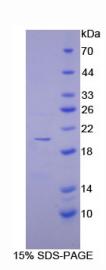
Organism Species: Homo sapiens (Human)
Instruction manual
FOR IN VITRO USE AND RESEARCH USE ONLY
NOT FOR USE IN CLINICAL DIAGNOSTIC PROCEDURES
9th Edition (Revised in Jul, 2013)
精神分裂症破坏蛋白1(DISC1)重组蛋白[ PROPERTIES ]
Residues: Ser102~Cys261 (Accession # Q9NRI5),
with two N-terminal Tags, His-tag and T7-tag. Host: E. coli
Subcellular Location: Cytoplasm. Cytoskeleton,
microtubule organizing center, centrosome. Cell
junction, synapse, postsynaptic cell membrane,
postsynaptic density.
Purity: >95%
Endotoxin Level: <1.0EU per 1μg
(determined by the LAL method).
精神分裂症破坏蛋白1(DISC1)重组蛋白Formulation: Supplied as lyophilized form in PBS,
pH7.4, containing 5% sucrose, 0.01% sarcosyl.
Predicted isoelectric point: 6.1
Predicted Molecular Mass: 20.2kDa
Applications: SDS-PAGE; WB; ELISA; IP.
(May be suitable for use in other assays to be determined by the end user.)
[ USAGE ]
Reconstitute in sterile PBS, pH7.2-pH7.4.
[ STORAGE AND STABILITY ]
Storage: Avoid repeated freeze/thaw cycles.
Store at 2-8oC for one month.
Aliquot and store at -80oC for 12 months.
Stability Test: The thermal stability is described by the loss rate of the target
protein. The loss rate was determined by accelerated thermal degradation test,
that is, incubate the protein at 37oC for 48h, and no obvious degradation and
精神分裂症破坏蛋白1(DISC1)重组蛋白precipitation were observed. (Referring from China Biological Products Standard,
which was calculated by the Arrhenius equation.) The loss of this protein is less
than 5% within the expiration date under appropriate storage condition.
[ SEQUENCES ]
The target protein is fused with two N-terminal Tags, His-tag and T7-tag, its
sequence is listed below.
MGSSHHHHHH SSGLVPRGSH MASMTGGQQM GRGSEF- SAAAPTVTS VRGTSAHFGI
QLRGGTRLP D RLSWP CGPGS AGWQQEFAAM DSSETLDASW EAACS DGARR
VRAAGSLPSA ELSSNSCSPG CGPEVPPTPP GSHSAFTSSF SFIRLSLGSA GERGEAEGCP
PSREAESHCQ SPQEMGAKAA SLDGPHEDPR C
